%matplotlib inline
How to “get started”¶
In this introductory example, you will see how to use SpikeInterface to perform a full electrophysiology analysis. We will download a simulated dataset, and we will then perform some pre-processing, run a spike sorting algorithm, post-process the spike sorting output, perform curation (manual and automatic), and compare spike sorting results.
import matplotlib.pyplot as plt
from pprint import pprint
The spikeinterface module by itself imports only the spikeinterface.core submodule which is not useful for the end user
import spikeinterface
We need to import one by one different submodules separately (preferred). There are several modules:
extractors: file IOpreprocessing: preprocessingsorters: Python wrappers of spike sorterspostprocessing: postprocessingqualitymetrics: quality metrics on units found by sorterscuration: automatic curation of spike sorting outputcomparison: comparison of spike sorting outputswidgets: visualization
import spikeinterface as si # import core only
import spikeinterface.extractors as se
import spikeinterface.preprocessing as spre
import spikeinterface.sorters as ss
import spikeinterface.postprocessing as spost
import spikeinterface.qualitymetrics as sqm
import spikeinterface.comparison as sc
import spikeinterface.exporters as sexp
import spikeinterface.curation as scur
import spikeinterface.widgets as sw
Alternatively, we can import all submodules at once with
import spikeinterface.full as si which internally imports
core+extractors+preprocessing+sorters+postprocessing+
qualitymetrics+comparison+widgets+exporters. In this case all aliases in
the following tutorial would be si.
This is useful for notebooks, but it is a heavier import because internally many more dependencies are imported (scipy/sklearn/networkx/matplotlib/h5py…)
import spikeinterface.full as si
Before getting started, we can set some global arguments for parallel processing. For this example, let’s use 4 jobs and time chunks of 1s:
global_job_kwargs = dict(n_jobs=4, chunk_duration="1s")
si.set_global_job_kwargs(**global_job_kwargs)
First, let’s download a simulated dataset from the https://gin.g-node.org/NeuralEnsemble/ephy_testing_data repo We download the dataset using DataLad but it can also be downloaded directly.
Then we can open it. Note that MEArec simulated files contain both a “recording” and a “sorting” object.
local_path = si.download_dataset(remote_path="mearec/mearec_test_10s.h5")
recording, sorting_true = se.read_mearec(local_path)
print(recording)
print(sorting_true)
MEArecRecordingExtractor: 32 channels - 32.0kHz - 1 segments - 320,000 samples - 10.00s
float32 dtype - 39.06 MiB
file_path: /home/nolanlab/spikeinterface_datasets/ephy_testing_data/mearec/mearec_test_10s.h5
MEArecSortingExtractor: 10 units - 1 segments - 32.0kHz
file_path: /home/nolanlab/spikeinterface_datasets/ephy_testing_data/mearec/mearec_test_10s.h5
recording is a BaseRecording object, which extracts information
about channel ids, channel locations (if present), the sampling
frequency of the recording, and the extracellular traces.
sorting_true is a BaseSorting object, which contains information
about spike-sorting related information, including unit ids, spike
trains, etc. Since the data are simulated, sorting_true has
ground-truth information of the spiking activity of each unit.
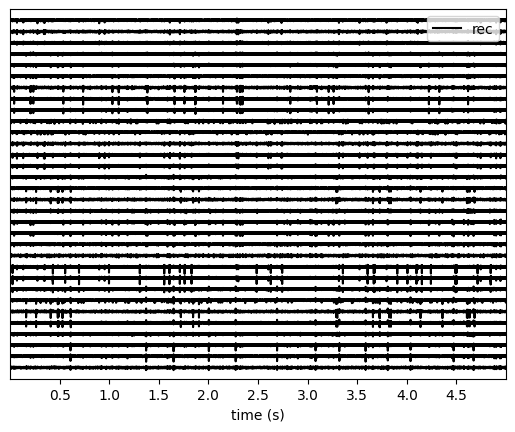
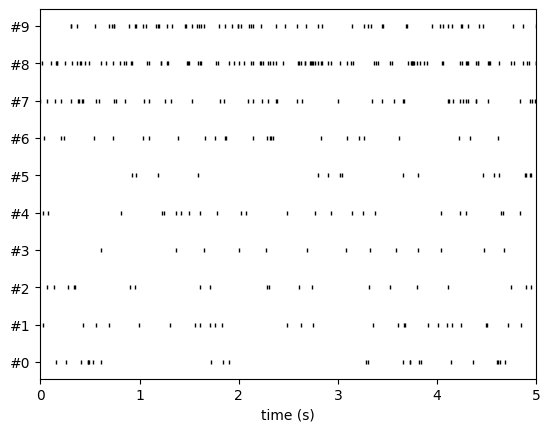
Let’s use the spikeinterface.widgets module to visualize the traces
and the raster plots.
w_ts = sw.plot_traces(recording, time_range=(0, 5))
w_rs = sw.plot_rasters(sorting_true, time_range=(0, 5))
This is how you retrieve info from a BaseRecording…
channel_ids = recording.get_channel_ids()
fs = recording.get_sampling_frequency()
num_chan = recording.get_num_channels()
num_seg = recording.get_num_segments()
print("Channel ids:", channel_ids)
print("Sampling frequency:", fs)
print("Number of channels:", num_chan)
print("Number of segments:", num_seg)
Channel ids: ['1' '2' '3' '4' '5' '6' '7' '8' '9' '10' '11' '12' '13' '14' '15' '16'
'17' '18' '19' '20' '21' '22' '23' '24' '25' '26' '27' '28' '29' '30'
'31' '32']
Sampling frequency: 32000.0
Number of channels: 32
Number of segments: 1
…and from a BaseSorting
num_seg = recording.get_num_segments()
unit_ids = sorting_true.get_unit_ids()
spike_train = sorting_true.get_unit_spike_train(unit_id=unit_ids[0])
print("Number of segments:", num_seg)
print("Unit ids:", unit_ids)
print("Spike train of first unit:", spike_train)
Number of segments: 1
Unit ids: ['#0' '#1' '#2' '#3' '#4' '#5' '#6' '#7' '#8' '#9']
Spike train of first unit: [ 5197 8413 13124 15420 15497 15668 16929 19607 55107 59060
60958 105193 105569 117082 119243 119326 122293 122877 132413 139498
147402 147682 148271 149857 165454 170569 174319 176237 183598 192278
201535 217193 219715 221226 222967 223897 225338 243206 243775 248754
253184 253308 265132 266197 266662 283149 284716 287592 304025 305286
310438 310775 318460]
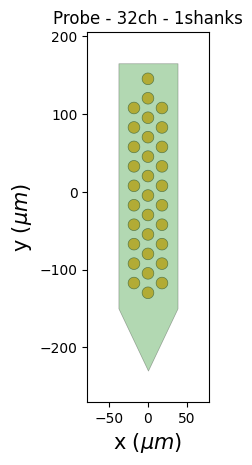
SpikeInterface internally uses the
ProbeInterface
package to handle probeinterface.Probe and
probeinterface.ProbeGroup. So any probe in the probeinterface
collection can be downloaded and set to a Recording object. In this
case, the MEArec dataset already handles a Probe and we don’t need
to set it manually.
probe = recording.get_probe()
print(probe)
from probeinterface.plotting import plot_probe
_ = plot_probe(probe)
Probe - 32ch - 1shanks
If your recording does not have a Probe, you can set it using
set_probe. Note: set_probe creates a copy of the recording with
the new probe, rather than modifying the existing recording in place.
There is more information
here.
Using the spikeinterface.preprocessing module, you can perform
preprocessing on the recordings. Each pre-processing function also
returns a BaseRecording, which makes it easy to build pipelines.
Here, we filter the recording and apply common median reference (CMR).
All these preprocessing steps are “lazy”. The computation is done on
demand when we call recording.get_traces(...) or when we save the
object to disk.
recording_cmr = recording
recording_f = si.bandpass_filter(recording, freq_min=300, freq_max=6000)
print(recording_f)
recording_cmr = si.common_reference(recording_f, reference="global", operator="median")
print(recording_cmr)
# this computes and saves the recording after applying the preprocessing chain
recording_preprocessed = recording_cmr.save(format="binary")
print(recording_preprocessed)
BandpassFilterRecording: 32 channels - 32.0kHz - 1 segments - 320,000 samples - 10.00s
float32 dtype - 39.06 MiB
CommonReferenceRecording: 32 channels - 32.0kHz - 1 segments - 320,000 samples - 10.00s
float32 dtype - 39.06 MiB
Use cache_folder=/tmp/spikeinterface_cache/tmpsgoh2z3y/DLGW1J8V
write_binary_recording with n_jobs = 4 and chunk_size = 32000
write_binary_recording: 0%| | 0/10 [00:00<?, ?it/s]
BinaryFolderRecording: 32 channels - 32.0kHz - 1 segments - 320,000 samples - 10.00s
float32 dtype - 39.06 MiB
To reload a preprocessed recording that was saved to disk, you can use load_extractor() function from the core module.
Now you are ready to spike sort using the spikeinterface.sorters
module! Let’s first check which sorters are implemented and which are
installed
print("Available sorters", ss.available_sorters())
print("Installed sorters", ss.installed_sorters())
Available sorters ['combinato', 'hdsort', 'herdingspikes', 'ironclust', 'kilosort', 'kilosort2', 'kilosort2_5', 'kilosort3', 'kilosort4', 'klusta', 'mountainsort4', 'mountainsort5', 'pykilosort', 'simple', 'spykingcircus', 'spykingcircus2', 'tridesclous', 'tridesclous2', 'waveclus', 'waveclus_snippets', 'yass']
Installed sorters ['herdingspikes', 'simple', 'spykingcircus2', 'tridesclous', 'tridesclous2']
The ss.installed_sorters() will list the sorters installed on the
machine. We can see we have HerdingSpikes and Tridesclous installed.
Spike sorters come with a set of parameters that users can change. The
available parameters are dictionaries and can be accessed with:
print("Tridesclous params:")
pprint(ss.get_default_sorter_params("tridesclous"))
print("SpykingCircus2 params:")
pprint(ss.get_default_sorter_params("spykingcircus2"))
Tridesclous params:
{'chunk_duration': '1s',
'common_ref_removal': False,
'detect_sign': -1,
'detect_threshold': 5,
'freq_max': 5000.0,
'freq_min': 400.0,
'max_threads_per_process': 1,
'mp_context': None,
'n_jobs': 20,
'nested_params': None,
'progress_bar': True}
SpykingCircus2 params:
{'apply_preprocessing': True,
'cache_preprocessing': {'delete_cache': True,
'memory_limit': 0.5,
'mode': None},
'clustering': {'legacy': False},
'debug': False,
'detection': {'detect_threshold': 4, 'peak_sign': 'neg'},
'filtering': {'freq_min': 150},
'general': {'ms_after': 2, 'ms_before': 2, 'radius_um': 100},
'job_kwargs': {'n_jobs': 0.8},
'matching': {'method': 'circus-omp-svd'},
'multi_units_only': False,
'selection': {'method': 'smart_sampling_amplitudes',
'min_n_peaks': 100000,
'n_peaks_per_channel': 5000,
'select_per_channel': False},
'sparsity': {'method': 'ptp', 'threshold': 0.25}}
Let’s run tridesclous and change one of the parameters, say, the
detect_threshold:
sorting_TDC = ss.run_sorter(sorter_name="tridesclous", recording=recording_preprocessed, detect_threshold=4)
print(sorting_TDC)
TridesclousSortingExtractor: 10 units - 1 segments - 32.0kHz
Alternatively we can pass a full dictionary containing the parameters:
other_params = ss.get_default_sorter_params("tridesclous")
other_params["detect_threshold"] = 6
# parameters set by params dictionary
sorting_TDC_2 = ss.run_sorter(
sorter_name="tridesclous", recording=recording_preprocessed, output_folder="tdc_output2", **other_params
)
print(sorting_TDC_2)
TridesclousSortingExtractor: 9 units - 1 segments - 32.0kHz
Let’s run spykingcircus2 as well, with default parameters:
sorting_SC2 = ss.run_sorter(sorter_name="spykingcircus2", recording=recording_preprocessed)
print(sorting_SC2)
NumpyFolderSorting: 10 units - 1 segments - 32.0kHz
The sorting_TDC and sorting_SC2 are BaseSorting objects. We
can print the units found using:
print("Units found by tridesclous:", sorting_TDC.get_unit_ids())
print("Units found by spyking-circus2:", sorting_SC2.get_unit_ids())
Units found by tridesclous: [0 1 2 3 4 5 6 7 8 9]
Units found by spyking-circus2: [0 1 2 3 4 5 6 7 8 9]
If a sorter is not installed locally, we can also avoid installing it
and run it anyways, using a container (Docker or Singularity). To do
this, you will need to install Docker. More information
here.
Let’s run Kilosort2 using Docker:
sorting_KS2 = ss.run_sorter(sorter_name="kilosort2", recording=recording_preprocessed, docker_image=True, verbose=True)
print(sorting_KS2)
installation_mode='auto' switching to installation_mode: 'dev'
Starting container
Installing spikeinterface with folder in container
Installing neo with pypi in container
Installing mearec with pypi in container
Running kilosort2 sorter inside spikeinterface/kilosort2-compiled-base
Stopping container
KiloSortSortingExtractor: 19 units - 1 segments - 32.0kHz
For postprocessing SpikeInterface pairs recording and sorting objects
into a SortingAnalyzer object. The SortingAnalyzer can be loaded
in memory or saved in a folder. Here, we save it in binary format.
analyzer_TDC = si.create_sorting_analyzer(sorting=sorting_TDC, recording=recording_preprocessed, format='binary_folder', folder='analyzer_TDC_binary')
estimate_sparsity: 0%| | 0/10 [00:00<?, ?it/s]
This folder is where all the postprocessing data will be saved such as
waveforms and templates. Let’s calculate some waveforms. When doing
this, the function samples some spikes (by default
max_spikes_per_unit=500) for each unit, extracts their waveforms,
and stores them to disk in
./analyzer_TDC_binary/extensions/waveforms. These waveforms are
helpful to compute the average waveform, or “template”, for each unit
and then to compute, for example, quality metrics. Computations with the
SortingAnalyzer object are done using the compute method:
analyzer_TDC.compute("random_spikes")
analyzer_TDC.compute("waveforms")
compute_waveforms: 0%| | 0/10 [00:00<?, ?it/s]
<spikeinterface.core.analyzer_extension_core.ComputeWaveforms at 0x7f49d5ddabb0>
The results of these calculations are saved as extensions. Some
simple data, such as the unit_ids can be accessed directly from the
SortingAnalyzer object. Extension data is accessed by first getting
the extension then getting the data
unit_id0 = analyzer_TDC.unit_ids[0]
waveforms = analyzer_TDC.get_extension("waveforms").get_data()[unit_id0]
print(waveforms.shape)
(96, 25)
There are many more properties we can calculate
analyzer_TDC.compute("noise_levels")
analyzer_TDC.compute("templates")
analyzer_TDC.compute("spike_amplitudes")
spike_amplitudes: 0%| | 0/10 [00:00<?, ?it/s]
<spikeinterface.postprocessing.spike_amplitudes.ComputeSpikeAmplitudes at 0x7f49d5e36340>
Many of the extensions have parameters you can tune
analyzer_TDC.compute("unit_locations", method="center_of_mass")
analyzer_TDC.compute("spike_locations", ms_before=0.5)
analyzer_TDC.compute("correlograms", bin_ms=0.1)
analyzer_TDC.compute("template_similarity", method="cosine_similarity")
spike_locations: 0%| | 0/10 [00:00<?, ?it/s]
<spikeinterface.postprocessing.template_similarity.ComputeTemplateSimilarity at 0x7f49d5e0ed90>
Find out more about the available parameters and extensions here.
The calculations are saved in the extensions subfolder of the
SortingAnalyzer folder. Similar to the waveforms we can access them
using get_extension and get_data. For example, here we can make
a historgram of spike amplitudes
amplitudes = analyzer_TDC.get_extension("spike_amplitudes").get_data()
plt.hist(amplitudes, bins=50)
plt.show()

You can check which extensions have been saved (in your local folder) and which have been loaded (in your enviroment)…
print(analyzer_TDC.get_saved_extension_names())
print(analyzer_TDC.get_loaded_extension_names())
['random_spikes', 'waveforms', 'templates', 'noise_levels', 'template_similarity', 'spike_amplitudes', 'correlograms', 'spike_locations', 'unit_locations']
['random_spikes', 'waveforms', 'noise_levels', 'templates', 'spike_amplitudes', 'unit_locations', 'spike_locations', 'correlograms', 'template_similarity']
…or delete an extension…
analyzer_TDC.delete_extension("spike_amplitudes")
This deletes the extension’s data in the SortingAnalyzer folder.
Importantly, SortingAnalyzers (and all extensions) can be reloaded
at later times from their folders: (Here, spike_amplitudes is not loaded
since we just deleted it)
sorting_analyzer_path = './analyzer_TDC_binary'
analyzer_loaded = si.load_sorting_analyzer(sorting_analyzer_path)
print(analyzer_loaded.get_loaded_extension_names())
['random_spikes', 'waveforms', 'templates', 'noise_levels', 'template_similarity', 'correlograms', 'spike_locations', 'unit_locations']
And any deleted extensions are easily recomputed
analyzer_TDC.compute("spike_amplitudes")
spike_amplitudes: 0%| | 0/10 [00:00<?, ?it/s]
<spikeinterface.postprocessing.spike_amplitudes.ComputeSpikeAmplitudes at 0x7f49b80eadc0>
Once we have computed all of the postprocessing information, we can
compute quality metrics (some quality metrics require certain extensions
- e.g., drift metrics require spike_locations):
qm_params = sqm.get_default_qm_params()
pprint(qm_params)
{'amplitude_cutoff': {'amplitudes_bins_min_ratio': 5,
'histogram_smoothing_value': 3,
'num_histogram_bins': 100,
'peak_sign': 'neg'},
'amplitude_cv': {'amplitude_extension': 'spike_amplitudes',
'average_num_spikes_per_bin': 50,
'min_num_bins': 10,
'percentiles': (5, 95)},
'amplitude_median': {'peak_sign': 'neg'},
'drift': {'direction': 'y',
'interval_s': 60,
'min_num_bins': 2,
'min_spikes_per_interval': 100},
'firing_range': {'bin_size_s': 5, 'percentiles': (5, 95)},
'isi_violation': {'isi_threshold_ms': 1.5, 'min_isi_ms': 0},
'nearest_neighbor': {'max_spikes': 10000, 'n_neighbors': 5},
'nn_isolation': {'max_spikes': 10000,
'min_fr': 0.0,
'min_spikes': 10,
'n_components': 10,
'n_neighbors': 4,
'peak_sign': 'neg',
'radius_um': 100},
'nn_noise_overlap': {'max_spikes': 10000,
'min_fr': 0.0,
'min_spikes': 10,
'n_components': 10,
'n_neighbors': 4,
'peak_sign': 'neg',
'radius_um': 100},
'presence_ratio': {'bin_duration_s': 60, 'mean_fr_ratio_thresh': 0.0},
'rp_violation': {'censored_period_ms': 0.0, 'refractory_period_ms': 1.0},
'silhouette': {'method': ('simplified',)},
'sliding_rp_violation': {'bin_size_ms': 0.25,
'contamination_values': None,
'exclude_ref_period_below_ms': 0.5,
'max_ref_period_ms': 10,
'min_spikes': 0,
'window_size_s': 1},
'snr': {'peak_mode': 'extremum', 'peak_sign': 'neg'},
'synchrony': {'synchrony_sizes': (2, 4, 8)}}
Since the recording is very short, let’s change some parameters to accommodate the duration:
qm_params["presence_ratio"]["bin_duration_s"] = 1
qm_params["amplitude_cutoff"]["num_histogram_bins"] = 5
qm_params["drift"]["interval_s"] = 2
qm_params["drift"]["min_spikes_per_interval"] = 2
Quality metrics are extensions, so computations and data extraction work in the same way as earlier
analyzer_TDC.compute("quality_metrics", qm_params)
analyzer_TDC.get_extension("quality_metrics").get_data()
| amplitude_cutoff | amplitude_cv_median | amplitude_cv_range | amplitude_median | drift_ptp | drift_std | drift_mad | firing_range | firing_rate | isi_violations_ratio | ... | num_spikes | presence_ratio | rp_contamination | rp_violations | sd_ratio | sliding_rp_violation | snr | sync_spike_2 | sync_spike_4 | sync_spike_8 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | NaN | NaN | NaN | -306.199036 | NaN | NaN | NaN | 0.72 | 3.0 | 0.0 | ... | 30.0 | NaN | 0.0 | 0.0 | 1.536918 | NaN | 27.140698 | 0.0 | 0.0 | 0.0 |
| 1 | NaN | NaN | NaN | -273.444977 | NaN | NaN | NaN | 0.18 | 5.1 | 0.0 | ... | 51.0 | NaN | 0.0 | 0.0 | 1.311148 | NaN | 24.059540 | 0.0 | 0.0 | 0.0 |
| 2 | NaN | NaN | NaN | -269.204590 | NaN | NaN | NaN | 0.90 | 5.3 | 0.0 | ... | 53.0 | NaN | 0.0 | 0.0 | 2.016703 | NaN | 24.387525 | 0.0 | 0.0 | 0.0 |
| 3 | NaN | NaN | NaN | -311.545715 | NaN | NaN | NaN | 0.72 | 5.0 | 0.0 | ... | 50.0 | NaN | 0.0 | 0.0 | 2.011083 | NaN | 26.948630 | 0.0 | 0.0 | 0.0 |
| 4 | NaN | NaN | NaN | -106.953278 | NaN | NaN | NaN | 0.72 | 3.6 | 0.0 | ... | 36.0 | NaN | 0.0 | 0.0 | 0.680199 | NaN | 9.585650 | 0.0 | 0.0 | 0.0 |
| 5 | NaN | NaN | NaN | -150.833191 | NaN | NaN | NaN | 0.36 | 4.2 | 0.0 | ... | 42.0 | NaN | 0.0 | 0.0 | 0.965515 | NaN | 13.196613 | 0.0 | 0.0 | 0.0 |
| 6 | NaN | NaN | NaN | -90.358444 | NaN | NaN | NaN | 0.00 | 4.8 | 0.0 | ... | 48.0 | NaN | 0.0 | 0.0 | 1.177009 | NaN | 8.193233 | 0.0 | 0.0 | 0.0 |
| 7 | NaN | NaN | NaN | -102.491577 | NaN | NaN | NaN | 2.34 | 19.3 | 0.0 | ... | 193.0 | NaN | 0.0 | 0.0 | 0.973417 | 0.155 | 8.808388 | 0.0 | 0.0 | 0.0 |
| 8 | NaN | NaN | NaN | -127.252319 | NaN | NaN | NaN | 0.90 | 12.9 | 0.0 | ... | 129.0 | NaN | 0.0 | 0.0 | 0.949695 | 0.310 | 11.125336 | 0.0 | 0.0 | 0.0 |
| 9 | NaN | NaN | NaN | -97.207291 | NaN | NaN | NaN | 2.16 | 11.0 | 0.0 | ... | 110.0 | NaN | 0.0 | 0.0 | 1.021080 | 0.270 | 8.281832 | 0.0 | 0.0 | 0.0 |
10 rows × 21 columns
And since the quality metrics are extensions, they are saved
SortingAnalyzer folder.
Now, we can use some of the powerful tools for spike sorting visualization.
We can export a sorting summary and quality metrics plot using the
sortingview backend. This will generate shareable links for
web-based visualization. For this to work you need to install
sortingview and construct a kachery-cloud:
https://github.com/magland/sortingview.
w1 = sw.plot_quality_metrics(analyzer_TDC, display=False, backend="sortingview")
w2 = sw.plot_sorting_summary(analyzer_TDC, display=False, curation=True, backend="sortingview")
The sorting summary plot can also be used for manual labeling and curation. In the example above, we manually merged two units (0, 4) and added accept labels (2, 6, 7). After applying our curation, we can click on the “Save as snapshot (sha://)” and copy the URI:
uri = "sha1://68cb54a9aaed2303fb82dedbc302c853e818f1b6"
sorting_curated_sv = scur.apply_sortingview_curation(sorting_TDC, uri_or_json=uri)
print(sorting_curated_sv)
print(sorting_curated_sv.get_property("accept"))
MergeUnitsSorting: 9 units - 1 segments - 32.0kHz
[False True False False True True False False False]
Alternatively, we can export the data locally to Phy. Phy is a GUI for manual curation of the spike sorting output. To export to phy you can run:
sexp.export_to_phy(analyzer_TDC, "phy_folder_for_TDC", verbose=True)
write_binary_recording: 0%| | 0/10 [00:00<?, ?it/s]
Fitting PCA: 0%| | 0/10 [00:00<?, ?it/s]
Projecting waveforms: 0%| | 0/10 [00:00<?, ?it/s]
extract PCs: 0%| | 0/10 [00:00<?, ?it/s]
Run:
phy template-gui /home/nolanlab/Chris/Developing/TestingDoc/phy_folder_for_TDC/params.py
Then you can run the template-gui with:
phy template-gui phy_folder_for_TDC/params.py and manually curate
the results.
After curating with Phy, the curated sorting can be reloaded to SpikeInterface. In this case, we exclude the units that have been labeled as “noise”:
sorting_curated_phy = se.read_phy("phy_folder_for_TDC", exclude_cluster_groups=["noise"])
Quality metrics can be also used to automatically curate the spike sorting output. For example, you can select sorted units with a SNR above a certain threshold:
qm_data = analyzer_TDC.get_extension("quality_metrics").get_data()
keep_mask = (qm_data["snr"] > 10) & (qm_data["isi_violations_ratio"] < 0.01)
print("Mask:", keep_mask.values)
sorting_curated_auto = sorting_TDC.select_units(sorting_TDC.unit_ids[keep_mask])
print(sorting_curated_auto)
Mask: [ True True True True False True False False True False]
UnitsSelectionSorting: 6 units - 1 segments - 32.0kHz
The final part of this tutorial deals with comparing spike sorting outputs. We can either:
compare the spike sorting results with the ground-truth sorting
sorting_truecompare the output of two sorters (e.g. Tridesclous and SpykingCircus2)
compare the output of multiple sorters (e.g. Tridesclous, SpykingCircus2, and Kilosort2)
comp_gt = sc.compare_sorter_to_ground_truth(gt_sorting=sorting_true, tested_sorting=sorting_TDC)
comp_pair = sc.compare_two_sorters(sorting1=sorting_TDC, sorting2=sorting_SC2)
comp_multi = sc.compare_multiple_sorters(
sorting_list=[sorting_TDC, sorting_SC2, sorting_KS2], name_list=["tdc", "sc2", "ks2"]
)
When comparing with a ground-truth sorting (1,), you can get the sorting performance and plot a confusion matrix
print(comp_gt.get_performance())
w_conf = sw.plot_confusion_matrix(comp_gt)
w_agr = sw.plot_agreement_matrix(comp_gt)
accuracy recall precision false_discovery_rate miss_rate
gt_unit_id
#0 1.0 1.0 1.0 0.0 0.0
#1 1.0 1.0 1.0 0.0 0.0
#2 0.976744 0.976744 1.0 0.0 0.023256
#3 1.0 1.0 1.0 0.0 0.0
#4 1.0 1.0 1.0 0.0 0.0
#5 0.972973 0.972973 1.0 0.0 0.027027
#6 1.0 1.0 1.0 0.0 0.0
#7 0.990991 0.990991 1.0 0.0 0.009009
#8 0.989744 0.989744 1.0 0.0 0.010256
#9 1.0 1.0 1.0 0.0 0.0


When comparing two sorters (2.), we can see the matching of units between sorters. Units which are not matched have -1 as their unit id:
comp_pair.hungarian_match_12
0 0.0
1 1.0
2 8.0
3 2.0
4 5.0
5 4.0
6 7.0
7 6.0
8 9.0
9 3.0
dtype: float64
or the reverse:
comp_pair.hungarian_match_21
0 0.0
1 1.0
2 3.0
3 9.0
4 5.0
5 4.0
6 7.0
7 6.0
8 2.0
9 8.0
dtype: float64
When comparing multiple sorters (3.), you can extract a BaseSorting
object with units in agreement between sorters. You can also plot a
graph showing how the units are matched between the sorters.
sorting_agreement = comp_multi.get_agreement_sorting(minimum_agreement_count=2)
print("Units in agreement between TDC, SC2, and KS2:", sorting_agreement.get_unit_ids())
w_multi = sw.plot_multicomparison_agreement(comp_multi)
w_multi = sw.plot_multicomparison_agreement_by_sorter(comp_multi)
Units in agreement between TDC, SC2, and KS2: [0 1 2 3 4 5 6 7 8 9]


We see that 10 unit were found by all sorters (note that this simulated dataset is a very simple example, and usually sorters do not do such a great job)!
However, Kilosort2 found 9 additional units that are not matched to ground-truth!
That’s all for this “How to get started” tutorial! Enjoy SpikeInterface!