%matplotlib inline
%load_ext autoreload
%autoreload 2
Handle motion/drift with spikeinterface¶
SpikeInterface offers a very flexible framework to handle drift as a preprocessing step. If you want to know more, please read the Motion/drift correction section of the documentation.
Here is a short demo on how to handle drift using the high-level
function correct_motion().
This function takes a preprocessed recording as input and then internally runs several steps (it can be slow!) and returns a lazy recording that interpolates the traces on-the-fly to compensate for the motion.
Internally this function runs the following steps:
1. localize_peaks()
2. select_peaks() (optional)
3. estimate_motion()
4. interpolate_motion()
All these sub-steps can be run with different methods and have many parameters.
The high-level function suggests 3 predifined “presets” and we will explore them using a very well known public dataset recorded by Nick Steinmetz: Imposed motion datasets
This dataset contains 3 recordings and each recording contains a Neuropixels 1 and a Neuropixels 2 probe.
Here we will use dataset1 with neuropixel1. This dataset is the “hello world” for drift correction in the spike sorting community!
from pathlib import Path
import matplotlib.pyplot as plt
import numpy as np
import shutil
import spikeinterface.full as si
base_folder = Path('/mnt/data/sam/DataSpikeSorting/imposed_motion_nick')
dataset_folder = base_folder / 'dataset1/NP1'
# read the file
raw_rec = si.read_spikeglx(dataset_folder)
raw_rec
SpikeGLXRecordingExtractor: 384 channels - 30.0kHz - 1 segments - 58,715,724 samples
1,957.19s (32.62 minutes) - int16 dtype - 42.00 GiB
We preprocess the recording with bandpass filter and a common median reference. Note, that it is better to not whiten the recording before motion estimation to get a better estimate of peak locations!
def preprocess_chain(rec):
rec = si.bandpass_filter(rec, freq_min=300., freq_max=6000.)
rec = si.common_reference(rec, reference='global', operator='median')
return rec
rec = preprocess_chain(raw_rec)
job_kwargs = dict(n_jobs=40, chunk_duration='1s', progress_bar=True)
Run motion correction with one function!¶
Correcting for drift is easy! You just need to run a single function. We will try this function with 3 presets.
Internally a preset is a dictionary of dictionaries containing all parameters for each step.
Here we also save the motion correction results into a folder to be able to load them later.
# internally, we can explore a preset like this
# every parameter can be overwritten at runtime
from spikeinterface.preprocessing.motion import motion_options_preset
motion_options_preset['kilosort_like']
{'doc': 'Mimic the drift correction of kilosrt (grid_convolution + iterative_template)',
'detect_kwargs': {'method': 'locally_exclusive',
'peak_sign': 'neg',
'detect_threshold': 8.0,
'exclude_sweep_ms': 0.1,
'radius_um': 50},
'select_kwargs': None,
'localize_peaks_kwargs': {'method': 'grid_convolution',
'radius_um': 30.0,
'upsampling_um': 3.0,
'sigma_um': array([ 5. , 12.5, 20. ]),
'sigma_ms': 0.25,
'margin_um': 30.0,
'prototype': None,
'percentile': 5.0},
'estimate_motion_kwargs': {'method': 'iterative_template',
'bin_duration_s': 2.0,
'rigid': False,
'win_step_um': 50.0,
'win_sigma_um': 150.0,
'margin_um': 0,
'win_shape': 'rect'},
'interpolate_motion_kwargs': {'direction': 1,
'border_mode': 'force_extrapolate',
'spatial_interpolation_method': 'kriging',
'sigma_um': [20.0, 30],
'p': 1}}
# lets try theses 3 presets
some_presets = ('rigid_fast', 'kilosort_like', 'nonrigid_accurate')
# some_presets = ('nonrigid_accurate', )
# compute motion with 3 presets
for preset in some_presets:
print('Computing with', preset)
folder = base_folder / 'motion_folder_dataset1' / preset
if folder.exists():
shutil.rmtree(folder)
recording_corrected, motion_info = si.correct_motion(rec, preset=preset,
folder=folder,
output_motion_info=True, **job_kwargs)
Plot the results¶
We load back the results and use the widgets module to explore the estimated drift motion.
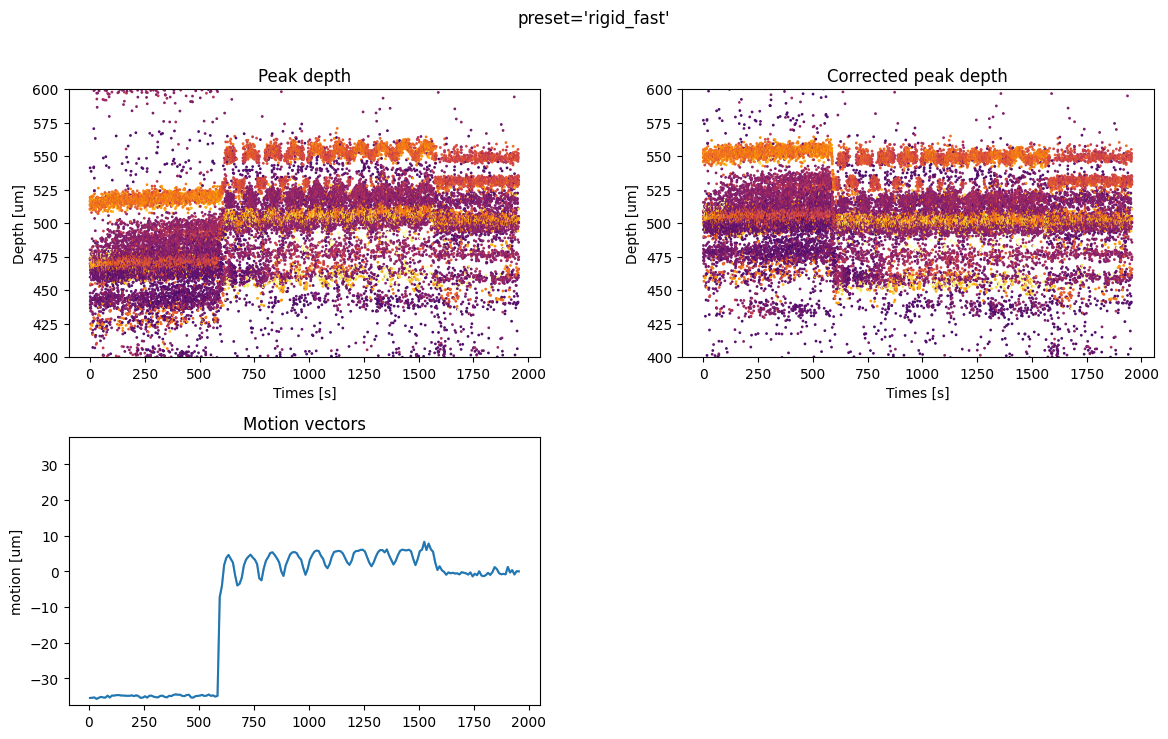
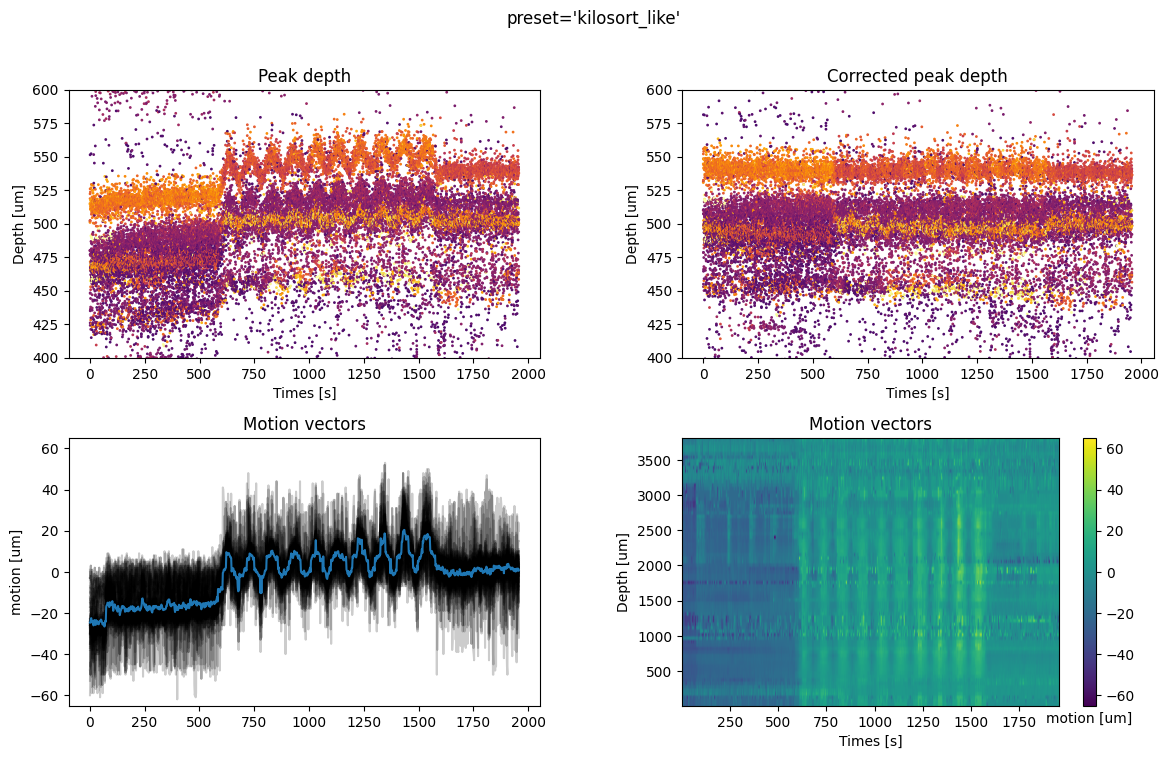
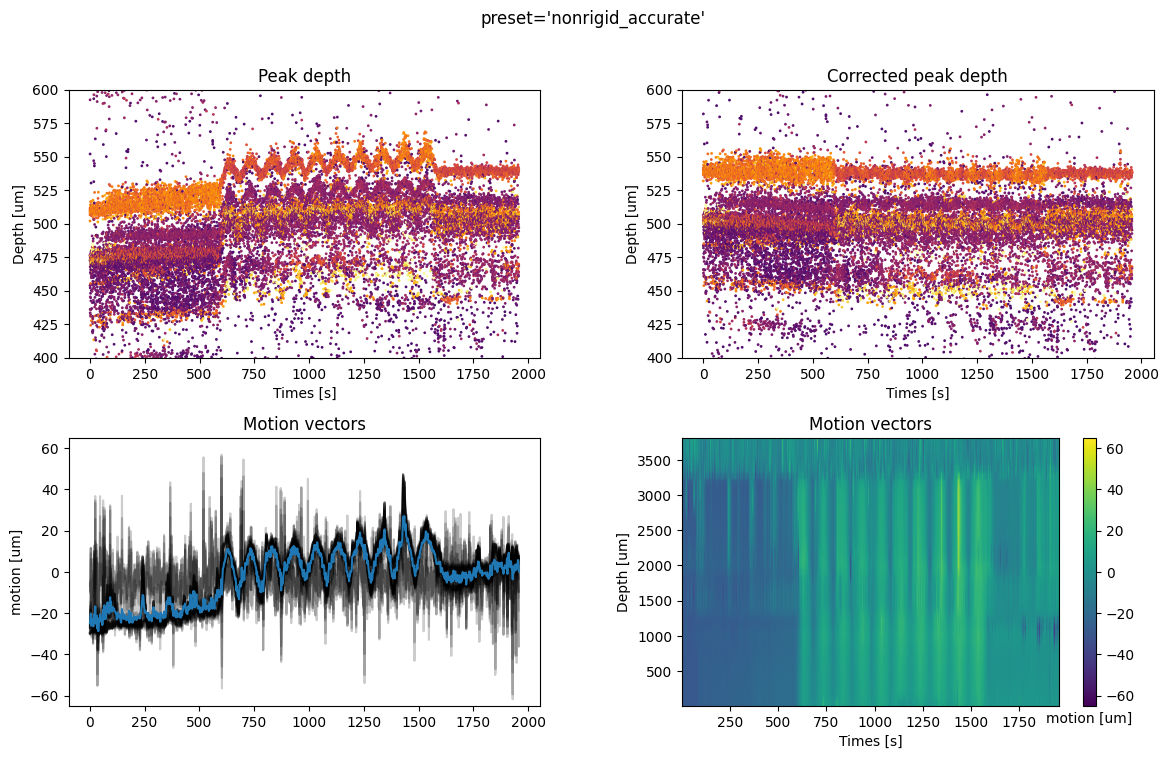
For all methods we have 4 plots:
top left: time vs estimated peak depth
top right: time vs peak depth after motion correction
bottom left: the average motion vector across depths and all motion across spatial depths (for non-rigid estimation)
bottom right: if motion correction is non rigid, the motion vector across depths is plotted as a map, with the color code representing the motion in micrometers.
- A few comments on the figures:
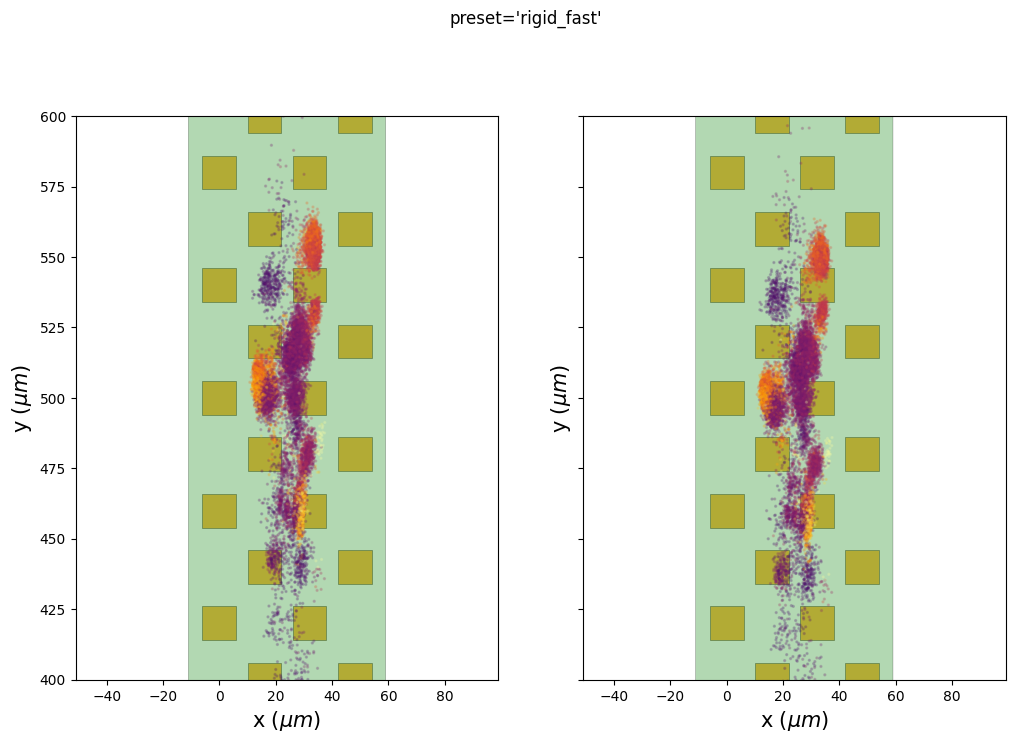
the preset ‘rigid_fast’ has only one motion vector for the entire probe because it is a “rigid” case. The motion amplitude is globally underestimated because it averages across depths. However, the corrected peaks are flatter than the non-corrected ones, so the job is partially done. The big jump at=600s when the probe start moving is recovered quite well.
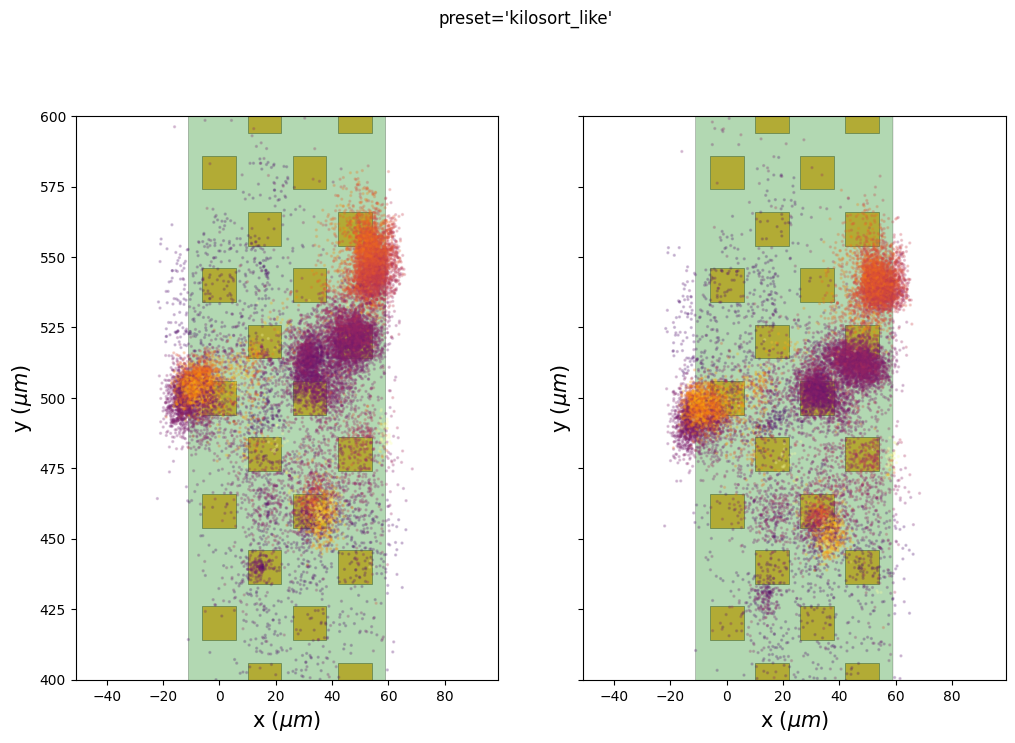
The preset kilosort_like gives better results because it is a non-rigid case. The motion vector is computed for different depths. The corrected peak locations are flatter than the rigid case. The motion vector map is still a bit noisy at some depths (e.g. around 1000um).
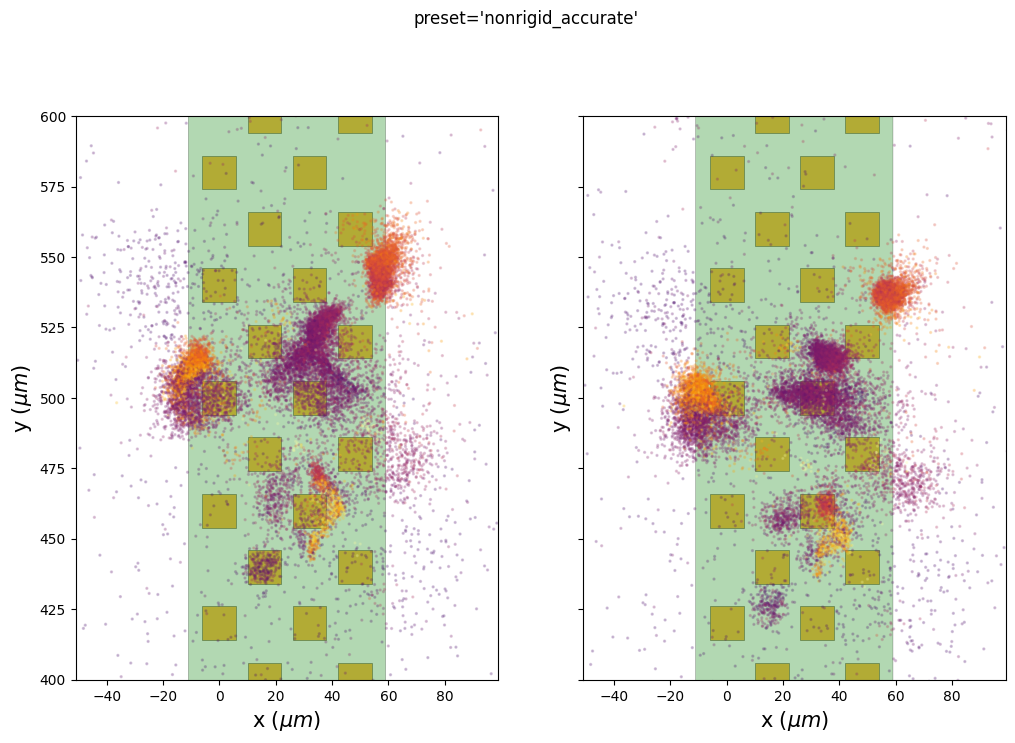
The preset nonrigid_accurate seems to give the best results on this recording. The motion vector seems less noisy globally, but it is not “perfect” (see at the top of the probe 3200um to 3800um). Also note that in the first part of the recording before the imposed motion (0-600s) we clearly have a non-rigid motion: the upper part of the probe (2000-3000um) experience some drift, but the lower part (0-1000um) is relatively stable. The method defined by this preset is able to capture this.
for preset in some_presets:
# load
folder = base_folder / 'motion_folder_dataset1' / preset
motion_info = si.load_motion_info(folder)
# and plot
fig = plt.figure(figsize=(14, 8))
si.plot_motion(motion_info, figure=fig, depth_lim=(400, 600),
color_amplitude=True, amplitude_cmap='inferno', scatter_decimate=10)
fig.suptitle(f"{preset=}")
Plot peak localization¶
We can also use the internal extra results (peaks and peaks location) to check if putative clusters have a lower spatial spread after the motion correction.
Here we plot the estimated peak locations (left) and the corrected peak locations (on right) on top of the probe. The color codes for the peak amplitudes.
We can see here that some clusters seem to be more compact on the ‘y’ axis, especially for the preset “nonrigid_accurate”.
Be aware that there are two ways to correct for the motion: 1.
Interpolate traces and detect/localize peaks again
(interpolate_recording()) 2. Compensate for drift directly on peak
locations (correct_motion_on_peaks())
Case 1 is used before running a spike sorter and the case 2 is used here to display the results.
from spikeinterface.sortingcomponents.motion_interpolation import correct_motion_on_peaks
for preset in some_presets:
folder = base_folder / 'motion_folder_dataset1' / preset
motion_info = si.load_motion_info(folder)
fig, axs = plt.subplots(ncols=2, figsize=(12, 8), sharey=True)
ax = axs[0]
si.plot_probe_map(rec, ax=ax)
peaks = motion_info['peaks']
sr = rec.get_sampling_frequency()
time_lim0 = 750.
time_lim1 = 1500.
mask = (peaks['sample_index'] > int(sr * time_lim0)) & (peaks['sample_index'] < int(sr * time_lim1))
sl = slice(None, None, 5)
amps = np.abs(peaks['amplitude'][mask][sl])
amps /= np.quantile(amps, 0.95)
c = plt.get_cmap('inferno')(amps)
color_kargs = dict(alpha=0.2, s=2, c=c)
loc = motion_info['peak_locations']
#color='black',
ax.scatter(loc['x'][mask][sl], loc['y'][mask][sl], **color_kargs)
loc2 = correct_motion_on_peaks(motion_info['peaks'], motion_info['peak_locations'], rec.sampling_frequency,
motion_info['motion'], motion_info['temporal_bins'], motion_info['spatial_bins'], direction="y")
ax = axs[1]
si.plot_probe_map(rec, ax=ax)
# color='black',
ax.scatter(loc2['x'][mask][sl], loc2['y'][mask][sl], **color_kargs)
ax.set_ylim(400, 600)
fig.suptitle(f"{preset=}")
run times¶
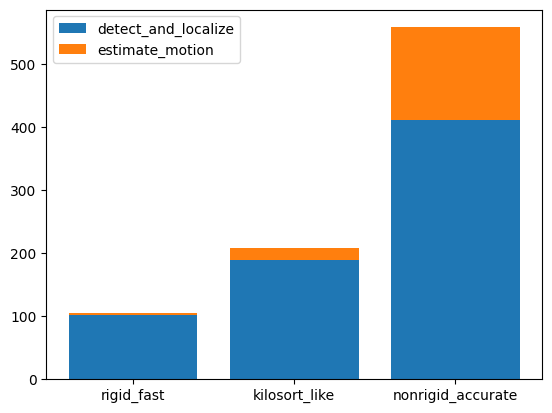
Presets and related methods have differents accuracies but also computation speeds. It is good to have this in mind!
run_times = []
for preset in some_presets:
folder = base_folder / 'motion_folder_dataset1' / preset
motion_info = si.load_motion_info(folder)
run_times.append(motion_info['run_times'])
keys = run_times[0].keys()
bottom = np.zeros(len(run_times))
fig, ax = plt.subplots()
for k in keys:
rtimes = np.array([rt[k] for rt in run_times])
if np.any(rtimes>0.):
ax.bar(some_presets, rtimes, bottom=bottom, label=k)
bottom += rtimes
ax.legend()
<matplotlib.legend.Legend at 0x7f0178a0ada0>