Note
Click here to download the full example code
Ground truth study tutorial¶
This tutorial illustrates how to run a “study”. A study is a systematic performance comparisons several ground truth recordings with several sorters.
The submodule study and the class propose high level tools functions to run many groundtruth comparison with many sorter on many recordings and then collect and aggregate results in an easy way.
The all mechanism is based on an intrinsic organization into a “study_folder” with several subfolder:
raw_files : contain a copy in binary format of recordings
sorter_folders : contains output of sorters
ground_truth : contains a copy of sorting ground in npz format
sortings: contains light copy of all sorting in npz format
tables: some table in cvs format
In order to run and rerun the computation all gt_sorting and recordings are copied to a fast and universal format : binary (for recordings) and npz (for sortings).
Imports
import matplotlib.pyplot as plt
import seaborn as sns
import spikeinterface.extractors as se
import spikeinterface.widgets as sw
from spikeinterface.comparison import GroundTruthStudy
Setup study folder and run all sorters¶
We first generate the folder. this can take some time because recordings are copied inside the folder.
rec0, gt_sorting0 = se.toy_example(num_channels=4, duration=10, seed=10, num_segments=1)
rec1, gt_sorting1 = se.toy_example(num_channels=4, duration=10, seed=0, num_segments=1)
gt_dict = {
'rec0': (rec0, gt_sorting0),
'rec1': (rec1, gt_sorting1),
}
study_folder = 'a_study_folder'
study = GroundTruthStudy.create(study_folder, gt_dict)
write_binary_recording with n_jobs = 1 and chunk_size = None
write_binary_recording with n_jobs = 1 and chunk_size = None
Then just run all sorters on all recordings in one functions.
# sorter_list = st.sorters.available_sorters() # this get all sorters.
sorter_list = ['herdingspikes', 'tridesclous', ]
study.run_sorters(sorter_list, mode_if_folder_exists="keep")
# Generating new position and neighbor files from data file
# Not Masking any Channels
# Sampling rate: 30000
# Localization On
# Number of recorded channels: 4
# Not subtracing mean
# Analysing frames: 300000; Seconds: 10.0
# Frames before spike in cutout: 9
# Frames after spike in cutout: 54
# tcuts: 39 84
# tInc: 100000
# Detection completed, time taken: 0:00:00.065673
# Time per frame: 0:00:00.000219
# Time per sample: 0:00:00.000055
Loaded 161 spikes.
Fitting dimensionality reduction using all spikes...
...projecting...
...done
Clustering...
Clustering 161 spikes...
number of seeds: 2
seeds/job: 2
using 2 cpus
[Parallel(n_jobs=2)]: Using backend LokyBackend with 2 concurrent workers.
[Parallel(n_jobs=2)]: Done 2 out of 2 | elapsed: 2.2s finished
/home/docs/checkouts/readthedocs.org/user_builds/spikeinterface/conda/0.95.0/lib/python3.8/site-packages/herdingspikes/clustering/mean_shift_.py:242: DeprecationWarning: `np.bool` is a deprecated alias for the builtin `bool`. To silence this warning, use `bool` by itself. Doing this will not modify any behavior and is safe. If you specifically wanted the numpy scalar type, use `np.bool_` here.
Deprecated in NumPy 1.20; for more details and guidance: https://numpy.org/devdocs/release/1.20.0-notes.html#deprecations
unique = np.ones(len(sorted_centers), dtype=np.bool)
/home/docs/checkouts/readthedocs.org/user_builds/spikeinterface/conda/0.95.0/lib/python3.8/site-packages/herdingspikes/clustering/mean_shift_.py:255: DeprecationWarning: `np.int` is a deprecated alias for the builtin `int`. To silence this warning, use `int` by itself. Doing this will not modify any behavior and is safe. When replacing `np.int`, you may wish to use e.g. `np.int64` or `np.int32` to specify the precision. If you wish to review your current use, check the release note link for additional information.
Deprecated in NumPy 1.20; for more details and guidance: https://numpy.org/devdocs/release/1.20.0-notes.html#deprecations
labels = np.zeros(n_samples, dtype=np.int)
Number of estimated units: 2
/home/docs/checkouts/readthedocs.org/user_builds/spikeinterface/conda/0.95.0/lib/python3.8/site-packages/tridesclous/dataio.py:175: DeprecationWarning: distutils Version classes are deprecated. Use packaging.version instead.
v1 = distutils.version.LooseVersion(tridesclous_version).version
/home/docs/checkouts/readthedocs.org/user_builds/spikeinterface/conda/0.95.0/lib/python3.8/site-packages/tridesclous/dataio.py:176: DeprecationWarning: distutils Version classes are deprecated. Use packaging.version instead.
v2 = distutils.version.LooseVersion(self.info['tridesclous_version']).version
# Generating new position and neighbor files from data file
# Not Masking any Channels
# Sampling rate: 30000
# Localization On
# Number of recorded channels: 4
# Not subtracing mean
# Analysing frames: 300000; Seconds: 10.0
# Frames before spike in cutout: 9
# Frames after spike in cutout: 54
# tcuts: 39 84
# tInc: 100000
# Detection completed, time taken: 0:00:00.067018
# Time per frame: 0:00:00.000223
# Time per sample: 0:00:00.000056
Loaded 163 spikes.
Fitting dimensionality reduction using all spikes...
...projecting...
...done
Clustering...
Clustering 163 spikes...
number of seeds: 2
seeds/job: 2
using 2 cpus
[Parallel(n_jobs=2)]: Using backend LokyBackend with 2 concurrent workers.
[Parallel(n_jobs=2)]: Done 2 out of 2 | elapsed: 2.2s finished
/home/docs/checkouts/readthedocs.org/user_builds/spikeinterface/conda/0.95.0/lib/python3.8/site-packages/herdingspikes/clustering/mean_shift_.py:242: DeprecationWarning: `np.bool` is a deprecated alias for the builtin `bool`. To silence this warning, use `bool` by itself. Doing this will not modify any behavior and is safe. If you specifically wanted the numpy scalar type, use `np.bool_` here.
Deprecated in NumPy 1.20; for more details and guidance: https://numpy.org/devdocs/release/1.20.0-notes.html#deprecations
unique = np.ones(len(sorted_centers), dtype=np.bool)
/home/docs/checkouts/readthedocs.org/user_builds/spikeinterface/conda/0.95.0/lib/python3.8/site-packages/herdingspikes/clustering/mean_shift_.py:255: DeprecationWarning: `np.int` is a deprecated alias for the builtin `int`. To silence this warning, use `int` by itself. Doing this will not modify any behavior and is safe. When replacing `np.int`, you may wish to use e.g. `np.int64` or `np.int32` to specify the precision. If you wish to review your current use, check the release note link for additional information.
Deprecated in NumPy 1.20; for more details and guidance: https://numpy.org/devdocs/release/1.20.0-notes.html#deprecations
labels = np.zeros(n_samples, dtype=np.int)
Number of estimated units: 2
/home/docs/checkouts/readthedocs.org/user_builds/spikeinterface/conda/0.95.0/lib/python3.8/site-packages/tridesclous/dataio.py:175: DeprecationWarning: distutils Version classes are deprecated. Use packaging.version instead.
v1 = distutils.version.LooseVersion(tridesclous_version).version
/home/docs/checkouts/readthedocs.org/user_builds/spikeinterface/conda/0.95.0/lib/python3.8/site-packages/tridesclous/dataio.py:176: DeprecationWarning: distutils Version classes are deprecated. Use packaging.version instead.
v2 = distutils.version.LooseVersion(self.info['tridesclous_version']).version
/home/docs/checkouts/readthedocs.org/user_builds/spikeinterface/conda/0.95.0/lib/python3.8/site-packages/tridesclous/dataio.py:175: DeprecationWarning: distutils Version classes are deprecated. Use packaging.version instead.
v1 = distutils.version.LooseVersion(tridesclous_version).version
/home/docs/checkouts/readthedocs.org/user_builds/spikeinterface/conda/0.95.0/lib/python3.8/site-packages/tridesclous/dataio.py:176: DeprecationWarning: distutils Version classes are deprecated. Use packaging.version instead.
v2 = distutils.version.LooseVersion(self.info['tridesclous_version']).version
You can re run run_study_sorters as many time as you want. By default mode=’keep’ so only uncomputed sorters are rerun. For instance, so just remove the “sorter_folders/rec1/herdingspikes” to re-run only one sorter on one recording.
Then we copy the spike sorting outputs into a separate subfolder. This allow to remove the “large” sorter_folders.
study.copy_sortings()
/home/docs/checkouts/readthedocs.org/user_builds/spikeinterface/conda/0.95.0/lib/python3.8/site-packages/tridesclous/dataio.py:175: DeprecationWarning: distutils Version classes are deprecated. Use packaging.version instead.
v1 = distutils.version.LooseVersion(tridesclous_version).version
/home/docs/checkouts/readthedocs.org/user_builds/spikeinterface/conda/0.95.0/lib/python3.8/site-packages/tridesclous/dataio.py:176: DeprecationWarning: distutils Version classes are deprecated. Use packaging.version instead.
v2 = distutils.version.LooseVersion(self.info['tridesclous_version']).version
Collect comparisons¶
You can collect in one shot all results and run the GroundTruthComparison on it. So you can access finely to all individual results.
Note that exhaustive_gt=True when you exactly how many units in ground truth (for synthetic datasets)
study.run_comparisons(exhaustive_gt=True)
for (rec_name, sorter_name), comp in study.comparisons.items():
print('*' * 10)
print(rec_name, sorter_name)
print(comp.count_score) # raw counting of tp/fp/...
comp.print_summary()
perf_unit = comp.get_performance(method='by_unit')
perf_avg = comp.get_performance(method='pooled_with_average')
m = comp.get_confusion_matrix()
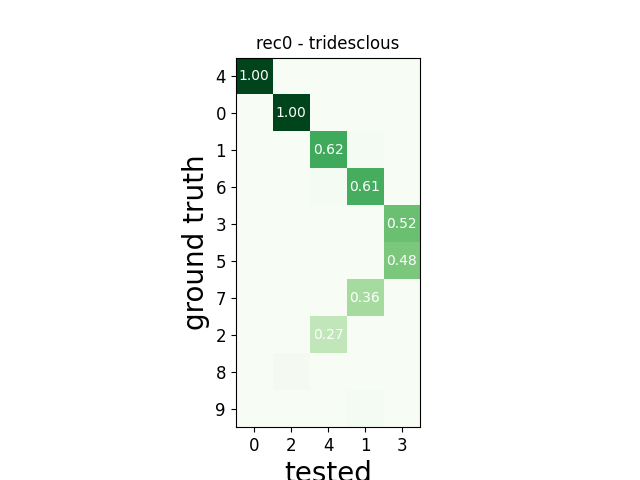
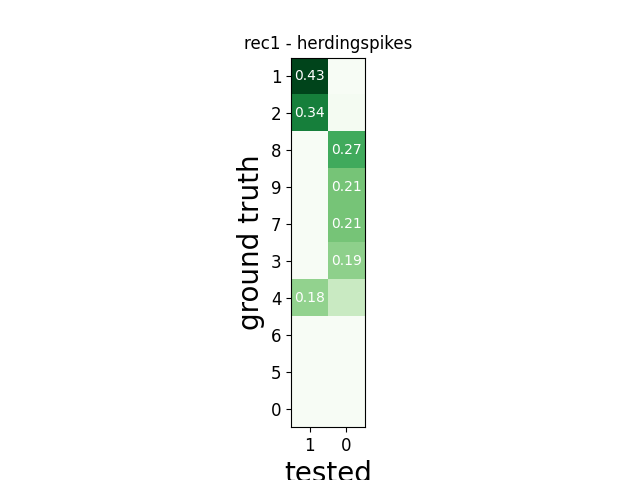
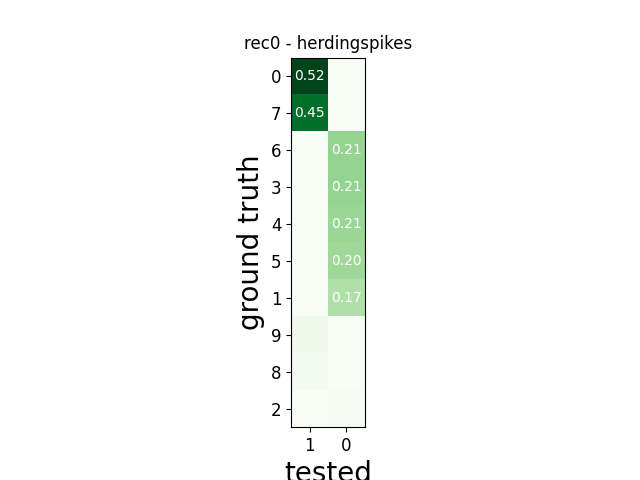
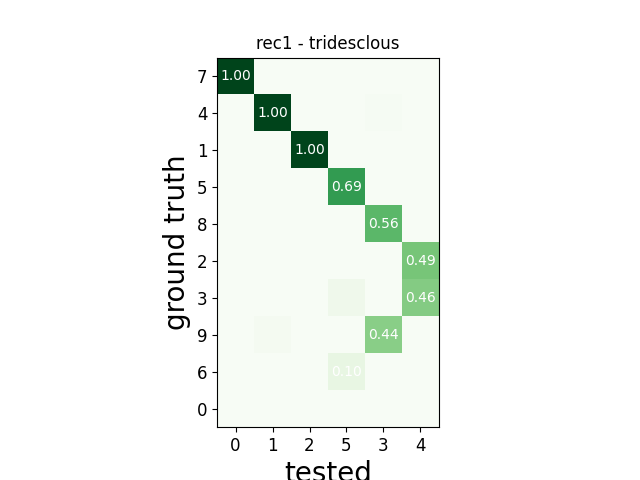
w_comp = sw.plot_agreement_matrix(comp)
w_comp.ax.set_title(rec_name + ' - ' + sorter_name)
**********
rec0 tridesclous
tp fn fp num_gt num_tested tested_id
gt_unit_id
0 23 0 0 23 23 2.0
1 20 0 12 20 32 4.0
2 0 24 0 24 0 -1.0
3 25 0 23 25 48 3.0
4 24 0 0 24 24 0.0
5 0 23 0 23 0 -1.0
6 25 0 16 25 41 1.0
7 0 20 0 20 0 -1.0
8 0 23 0 23 0 -1.0
9 0 22 0 22 0 -1.0
SUMMARY
-------
GT num_units: 10
TESTED num_units: 5
num_well_detected: 2
num_redundant: 0
num_overmerged: 3
num_false_positive_units 0
num_bad: 0
**********
rec1 herdingspikes
tp fn fp num_gt num_tested tested_id
gt_unit_id
0 0 22 0 22 0 -1.0
1 0 26 0 26 0 -1.0
2 0 22 0 22 0 -1.0
3 0 25 0 25 0 -1.0
4 0 25 0 25 0 -1.0
5 0 27 0 27 0 -1.0
6 0 22 0 22 0 -1.0
7 0 22 0 22 0 -1.0
8 0 28 0 28 0 -1.0
9 0 22 0 22 0 -1.0
SUMMARY
-------
GT num_units: 10
TESTED num_units: 2
num_well_detected: 0
num_redundant: 0
num_overmerged: 2
num_false_positive_units 0
num_bad: 2
**********
rec0 herdingspikes
tp fn fp num_gt num_tested tested_id
gt_unit_id
0 23 0 21 23 44 1.0
1 0 20 0 20 0 -1.0
2 0 24 0 24 0 -1.0
3 0 25 0 25 0 -1.0
4 0 24 0 24 0 -1.0
5 0 23 0 23 0 -1.0
6 0 25 0 25 0 -1.0
7 0 20 0 20 0 -1.0
8 0 23 0 23 0 -1.0
9 0 22 0 22 0 -1.0
SUMMARY
-------
GT num_units: 10
TESTED num_units: 2
num_well_detected: 0
num_redundant: 0
num_overmerged: 2
num_false_positive_units 0
num_bad: 1
**********
rec1 tridesclous
tp fn fp num_gt num_tested tested_id
gt_unit_id
0 0 22 0 22 0 -1.0
1 26 0 0 26 26 2.0
2 0 22 0 22 0 -1.0
3 0 25 0 25 0 -1.0
4 25 0 0 25 25 1.0
5 24 3 8 27 32 5.0
6 0 22 0 22 0 -1.0
7 22 0 0 22 22 0.0
8 28 0 22 28 50 3.0
9 0 22 0 22 0 -1.0
SUMMARY
-------
GT num_units: 10
TESTED num_units: 6
num_well_detected: 3
num_redundant: 0
num_overmerged: 2
num_false_positive_units 0
num_bad: 1
Collect synthetic dataframes and display¶
As shown previously, the performance is returned as a pandas dataframe.
The aggregate_performances_table() function, gathers all the outputs in
the study folder and merges them in a single dataframe.
dataframes = study.aggregate_dataframes()
Pandas dataframes can be nicely displayed as tables in the notebook.
print(dataframes.keys())
dict_keys(['run_times', 'perf_by_unit', 'count_units'])
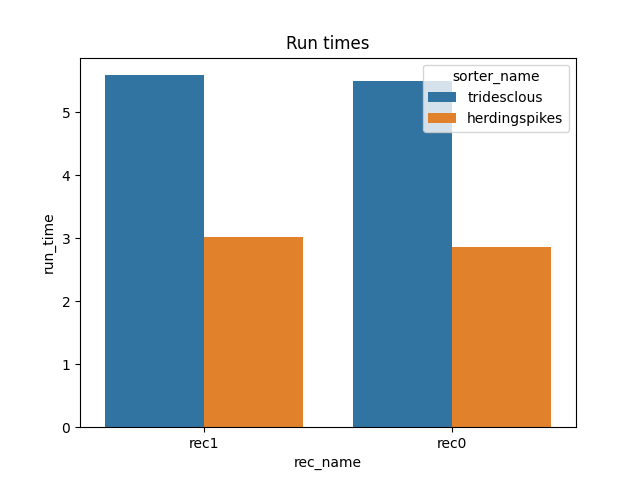
print(dataframes['run_times'])
rec_name sorter_name run_time
0 rec1 tridesclous 5.588428
1 rec0 tridesclous 5.495355
2 rec1 herdingspikes 3.022241
3 rec0 herdingspikes 2.862374
Easy plot with seaborn¶
Seaborn allows to easily plot pandas dataframes. Let’s see some examples.
run_times = dataframes['run_times']
fig1, ax1 = plt.subplots()
sns.barplot(data=run_times, x='rec_name', y='run_time', hue='sorter_name', ax=ax1)
ax1.set_title('Run times')
/home/docs/checkouts/readthedocs.org/user_builds/spikeinterface/conda/0.95.0/lib/python3.8/site-packages/seaborn/categorical.py:381: DeprecationWarning: distutils Version classes are deprecated. Use packaging.version instead.
if LooseVersion(mpl.__version__) < "3.0":
/home/docs/checkouts/readthedocs.org/user_builds/spikeinterface/conda/0.95.0/lib/python3.8/site-packages/setuptools/_distutils/version.py:346: DeprecationWarning: distutils Version classes are deprecated. Use packaging.version instead.
other = LooseVersion(other)
Text(0.5, 1.0, 'Run times')
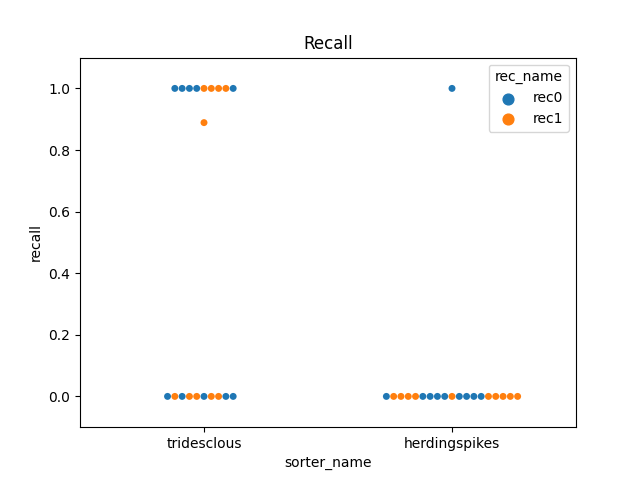
perfs = dataframes['perf_by_unit']
fig2, ax2 = plt.subplots()
sns.swarmplot(data=perfs, x='sorter_name', y='recall', hue='rec_name', ax=ax2)
ax2.set_title('Recall')
ax2.set_ylim(-0.1, 1.1)
plt.show()
/home/docs/checkouts/readthedocs.org/user_builds/spikeinterface/conda/0.95.0/lib/python3.8/site-packages/seaborn/categorical.py:381: DeprecationWarning: distutils Version classes are deprecated. Use packaging.version instead.
if LooseVersion(mpl.__version__) < "3.0":
/home/docs/checkouts/readthedocs.org/user_builds/spikeinterface/conda/0.95.0/lib/python3.8/site-packages/setuptools/_distutils/version.py:346: DeprecationWarning: distutils Version classes are deprecated. Use packaging.version instead.
other = LooseVersion(other)
Total running time of the script: ( 0 minutes 19.005 seconds)