Note
Click here to download the full example code
Preprocessing Tutorial¶
Before spike sorting, you may need to preproccess your signals in order to improve the spike sorting performance.
You can do that in SpikeInterface using the spikeinterface.preprocessing submodule.
import numpy as np
import matplotlib.pylab as plt
import scipy.signal
import spikeinterface.extractors as se
from spikeinterface.preprocessing import (bandpass_filter, notch_filter, common_reference,
remove_artifacts, preprocesser_dict)
First, let’s create a toy example:
recording, sorting = se.toy_example(num_channels=4, duration=10, seed=0)
Apply filters¶
Now apply a bandpass filter and a notch filter (separately) to the
recording extractor. Filters are also BaseRecording objects.
Note that these operation are lazy the computation is done on the fly
with rec.get_traces()
recording_bp = bandpass_filter(recording, freq_min=300, freq_max=6000)
print(recording_bp)
recording_notch = notch_filter(recording, freq=2000, q=30)
print(recording_notch)
BandpassFilterRecording: 4 channels - 2 segments - 30.0kHz - 20.000s
NotchFilterRecording: 4 channels - 2 segments - 30.0kHz - 20.000s
Now let’s plot the power spectrum of non-filtered, bandpass filtered, and notch filtered recordings.
fs = recording.get_sampling_frequency()
f_raw, p_raw = scipy.signal.welch(recording.get_traces(segment_index=0)[:, 0], fs=fs)
f_bp, p_bp = scipy.signal.welch(recording_bp.get_traces(segment_index=0)[:, 0], fs=fs)
f_notch, p_notch = scipy.signal.welch(recording_notch.get_traces(segment_index=0)[:, 0], fs=fs)
fig, ax = plt.subplots()
ax.semilogy(f_raw, p_raw, f_bp, p_bp, f_notch, p_notch)
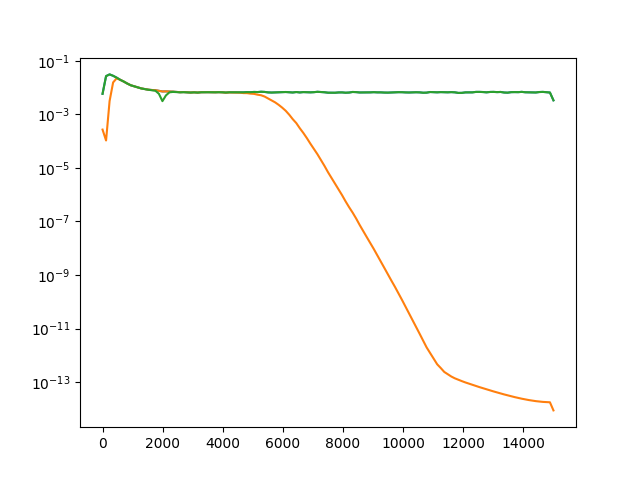
[<matplotlib.lines.Line2D object at 0x7f2978211430>, <matplotlib.lines.Line2D object at 0x7f2978211190>, <matplotlib.lines.Line2D object at 0x7f29782115e0>]
Change reference¶
In many cases, before spike sorting, it is wise to re-reference the signals to reduce the common-mode noise from the recordings.
To re-reference in spikeinterface.preprocessing you can use the
common_reference()
function. Both common average reference (CAR) and common median
reference (CMR) can be applied. Moreover, the average/median can be
computed on different groups. Single channels can also be used as
reference.
recording_car = common_reference(recording, reference='global', operator='average')
recording_cmr = common_reference(recording, reference='global', operator='median')
recording_single = common_reference(recording, reference='single', ref_channel_ids=[1])
recording_single_groups = common_reference(recording, reference='single',
groups=[[0, 1], [2, 3]],
ref_channel_ids=[0, 2])
trace0_car = recording_car.get_traces(segment_index=0)[:, 0]
trace0_cmr = recording_cmr.get_traces(segment_index=0)[:, 0]
trace0_single = recording_single.get_traces(segment_index=0)[:, 0]
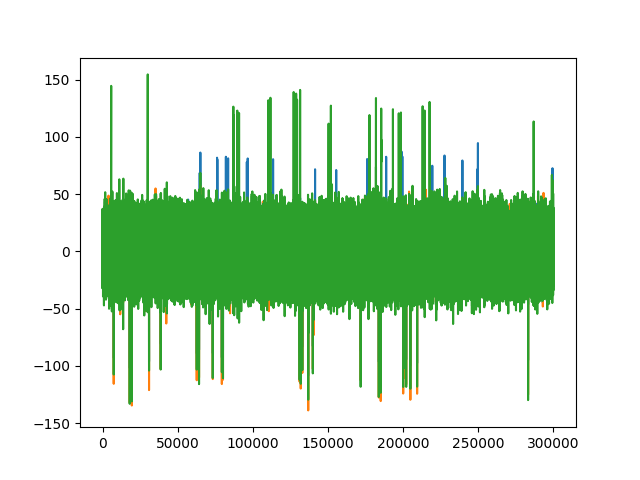
fig1, ax1 = plt.subplots()
ax1.plot(trace0_car)
ax1.plot(trace0_cmr)
ax1.plot(trace0_single)
trace1_groups = recording_single_groups.get_traces(segment_index=0)[:, 1]
trace0_groups = recording_single_groups.get_traces(segment_index=0)[:, 0]
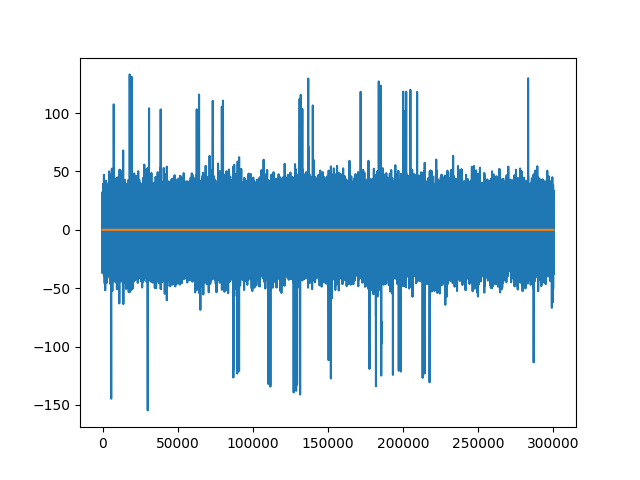
fig2, ax2 = plt.subplots()
ax2.plot(trace1_groups) # not zero
ax2.plot(trace0_groups)
[<matplotlib.lines.Line2D object at 0x7f297801e1c0>]
Remove stimulation artifacts¶
In some applications, electrodes are used to electrically stimulate the
tissue, generating a large artifact. In spikeinterface.preprocessing, the artifact
can be zeroed-out using the remove_artifacts() function.
# create dummy stimulation triggers per segment
stimulation_trigger_frames = [
[10000, 150000, 200000],
[20000, 30000],
]
# large ms_before and s_after are used for plotting only
recording_rm_artifact = remove_artifacts(recording, stimulation_trigger_frames,
ms_before=100, ms_after=200)
trace0 = recording.get_traces(segment_index=0)[:, 0]
trace0_rm = recording_rm_artifact.get_traces(segment_index=0)[:, 0]
fig3, ax3 = plt.subplots()
ax3.plot(trace0)
ax3.plot(trace0_rm)
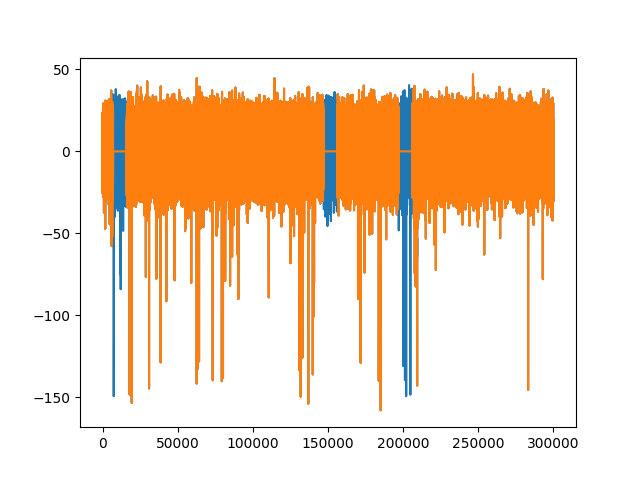
[<matplotlib.lines.Line2D object at 0x7f29b0a49130>]
You can list the available preprocessors with:
from pprint import pprint
pprint(preprocesser_dict)
plt.show()
{'bandpass_filter': <class 'spikeinterface.preprocessing.filter.BandpassFilterRecording'>,
'blank_staturation': <class 'spikeinterface.preprocessing.clip.BlankSaturationRecording'>,
'center': <class 'spikeinterface.preprocessing.normalize_scale.ZScoreRecording'>,
'clip': <class 'spikeinterface.preprocessing.clip.ClipRecording'>,
'common_reference': <class 'spikeinterface.preprocessing.common_reference.CommonReferenceRecording'>,
'deepinterpolate': <class 'spikeinterface.preprocessing.deepinterpolation.deepinterpolation.DeepInterpolatedRecording'>,
'filter': <class 'spikeinterface.preprocessing.filter.FilterRecording'>,
'highpass_filter': <class 'spikeinterface.preprocessing.filter.HighpassFilterRecording'>,
'normalize_by_quantile': <class 'spikeinterface.preprocessing.normalize_scale.NormalizeByQuantileRecording'>,
'notch_filter': <class 'spikeinterface.preprocessing.filter.NotchFilterRecording'>,
'phase_shift': <class 'spikeinterface.preprocessing.phase_shift.PhaseShiftRecording'>,
'rectify': <class 'spikeinterface.preprocessing.rectify.RectifyRecording'>,
'remove_artifacts': <class 'spikeinterface.preprocessing.remove_artifacts.RemoveArtifactsRecording'>,
'remove_bad_channels': <class 'spikeinterface.preprocessing.remove_bad_channels.RemoveBadChannelsRecording'>,
'resample': <class 'spikeinterface.preprocessing.resample.ResampleRecording'>,
'scale': <class 'spikeinterface.preprocessing.normalize_scale.ScaleRecording'>,
'whiten': <class 'spikeinterface.preprocessing.whiten.WhitenRecording'>,
'zero_channel_pad': <class 'spikeinterface.preprocessing.zero_channel_pad.ZeroChannelPaddedRecording'>}
Total running time of the script: ( 0 minutes 1.636 seconds)