Note
Go to the end to download the full example code
Read various format into SpikeInterface¶
SpikeInterface can read various formats of “recording” (traces) and “sorting” (spike train) data.
- Internally, to read different formats, SpikeInterface either uses:
a wrapper to neo rawio classes
or a direct implementation
Note that:
file formats contain a “recording”, a “sorting”, or “both”
file formats can be file-based (NWB, …) or folder based (SpikeGLX, OpenEphys, …)
In this example we demonstrate how to read different file formats into SI
import matplotlib.pyplot as plt
import spikeinterface.core as si
import spikeinterface.extractors as se
Let’s download some datasets in different formats from the ephy_testing_data repo:
MEArec: a simulator format which is hdf5-based. It contains both a “recording” and a “sorting” in the same file.
Spike2: file from spike2 devices. It contains “recording” information only.
spike2_file_path = si.download_dataset(remote_path="spike2/130322-1LY.smr")
print(spike2_file_path)
mearec_folder_path = si.download_dataset(remote_path="mearec/mearec_test_10s.h5")
print(mearec_folder_path)
/home/docs/spikeinterface_datasets/ephy_testing_data/spike2/130322-1LY.smr
/home/docs/spikeinterface_datasets/ephy_testing_data/mearec/mearec_test_10s.h5
Now that we have downloaded the files, let’s load them into SI.
The read_spike2() function returns one object,
a BaseRecording.
Note that internally this file contains 2 data streams (‘0’ and ‘1’), so we need to specify which one we
want to retrieve (‘0’ in our case).
the stream information can be retrieved by using the get_neo_streams() function.
stream_names, stream_ids = se.get_neo_streams("spike2", spike2_file_path)
print(stream_names)
print(stream_ids)
stream_id = stream_ids[0]
print("stream_id", stream_id)
recording = se.read_spike2(spike2_file_path, stream_id="0")
print(recording)
print(type(recording))
print(isinstance(recording, si.BaseRecording))
['Signal stream 0', 'Signal stream 1']
['0', '1']
stream_id 0
Spike2RecordingExtractor: 1 channels - 20.8kHz - 1 segments - 4,126,365 samples
198.07s (3.30 minutes) - int16 dtype - 7.87 MiB
file_path: /home/docs/spikeinterface_datasets/ephy_testing_data/spike2/130322-1LY.smr
<class 'spikeinterface.extractors.neoextractors.spike2.Spike2RecordingExtractor'>
True
The read_spike2`() function is equivalent to instantiating a
Spike2RecordingExtractor object:
recording = se.Spike2RecordingExtractor(spike2_file_path, stream_id="0")
print(recording)
Spike2RecordingExtractor: 1 channels - 20.8kHz - 1 segments - 4,126,365 samples
198.07s (3.30 minutes) - int16 dtype - 7.87 MiB
file_path: /home/docs/spikeinterface_datasets/ephy_testing_data/spike2/130322-1LY.smr
The read_mearec() function returns two objects,
a BaseRecording and a BaseSorting:
recording, sorting = se.read_mearec(mearec_folder_path)
print(recording)
print(type(recording))
print()
print(sorting)
print(type(sorting))
MEArecRecordingExtractor: 32 channels - 32.0kHz - 1 segments - 320,000 samples - 10.00s
float32 dtype - 39.06 MiB
file_path: /home/docs/spikeinterface_datasets/ephy_testing_data/mearec/mearec_test_10s.h5
<class 'spikeinterface.extractors.neoextractors.mearec.MEArecRecordingExtractor'>
MEArecSortingExtractor: 10 units - 1 segments - 32.0kHz
file_path: /home/docs/spikeinterface_datasets/ephy_testing_data/mearec/mearec_test_10s.h5
<class 'spikeinterface.extractors.neoextractors.mearec.MEArecSortingExtractor'>
The read_mearec() function is equivalent to:
recording = se.MEArecRecordingExtractor(mearec_folder_path)
sorting = se.MEArecSortingExtractor(mearec_folder_path)
SI objects (BaseRecording and BaseSorting)
can be plotted quickly with the spikeinterface.widgets submodule:
import spikeinterface.widgets as sw
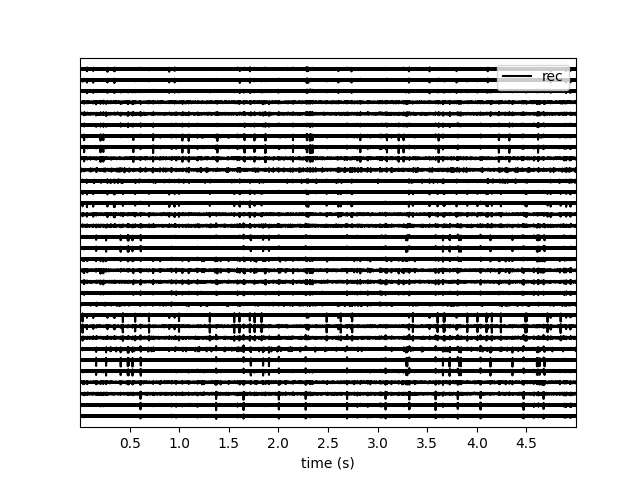
w_ts = sw.plot_traces(recording, time_range=(0, 5))
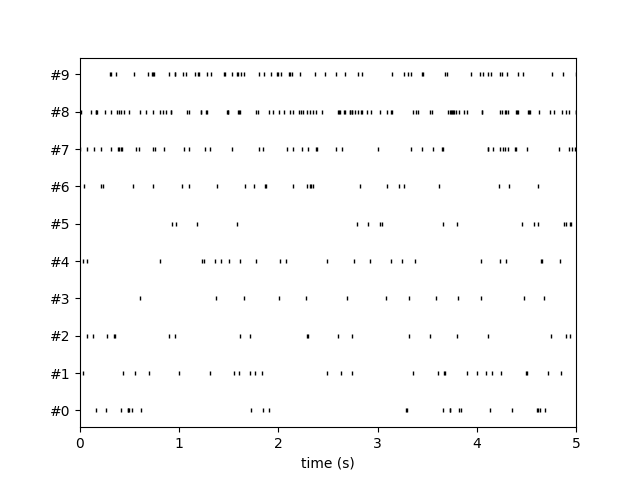
w_rs = sw.plot_rasters(sorting, time_range=(0, 5))
plt.show()
Total running time of the script: (0 minutes 44.235 seconds)