Note
Click here to download the full example code
Postprocessing Tutorial¶
Spike sorters generally output a set of units with corresponding spike trains. The toolkit.postprocessing
submodule allows to combine the RecordingExtractor and the sorted SortingExtractor objects to perform
further postprocessing.
import matplotlib.pylab as plt
import spikeinterface as si
import spikeinterface.extractors as se
import spikeinterface.toolkit as st
First, let’s download a simulated dataset from the repo ‘https://gin.g-node.org/NeuralEnsemble/ephy_testing_data’
local_path = si.download_dataset(remote_path='mearec/mearec_test_10s.h5')
recording = se.MEArecRecordingExtractor(local_path)
sorting = se.MEArecSortingExtractor(local_path)
print(recording)
print(sorting)
Out:
MEArecRecordingExtractor: 32 channels - 1 segments - 32.0kHz - 10.000s
file_path: /home/docs/spikeinterface_datasets/ephy_testing_data/mearec/mearec_test_10s.h5
MEArecSortingExtractor: 10 units - 1 segments - 32.0kHz
file_path: /home/docs/spikeinterface_datasets/ephy_testing_data/mearec/mearec_test_10s.h5
Assuming the sorting is the output of a spike sorter, the
postprocessing module allows to extract all relevant information
from the paired recording-sorting.
Compute spike waveforms¶
Waveforms are extracted with the WaveformExtractor or directly with the
extract_waveforms function (which returns a
WaveformExtractor object):
folder = 'waveforms_mearec'
we = si.extract_waveforms(recording, sorting, folder,
load_if_exists=True,
ms_before=1, ms_after=2., max_spikes_per_unit=500,
n_jobs=1, chunk_size=30000)
print(we)
Out:
WaveformExtractor: 32 channels - 10 units - 1 segments
before:32 after64 n_per_units: 500
Let’s plot the waveforms of units [0, 1, 2] on channel 8
colors = ['Olive', 'Teal', 'Fuchsia']
fig, ax = plt.subplots()
for i, unit_id in enumerate(sorting.unit_ids[:3]):
wf = we.get_waveforms(unit_id)
color = colors[i]
ax.plot(wf[:, :, 8].T, color=color, lw=0.3)
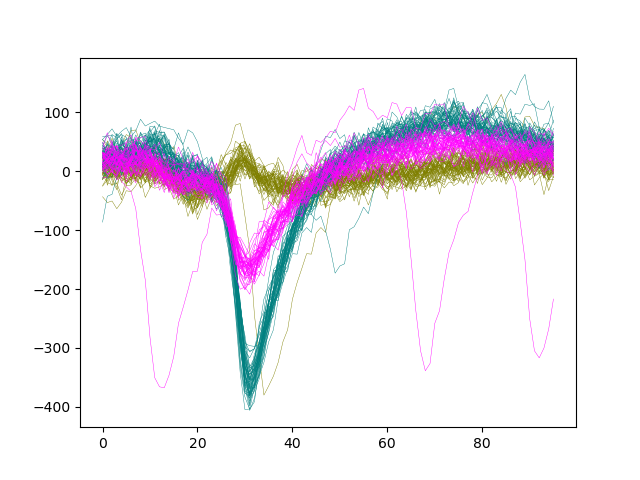
Compute unit templates¶
Similarly to waveforms, templates - average waveforms - can be easily retrieved
from the WaveformExtractor object:
fig, ax = plt.subplots()
for i, unit_id in enumerate(sorting.unit_ids[:3]):
template = we.get_template(unit_id)
color = colors[i]
ax.plot(template[:, 8].T, color=color, lw=3)
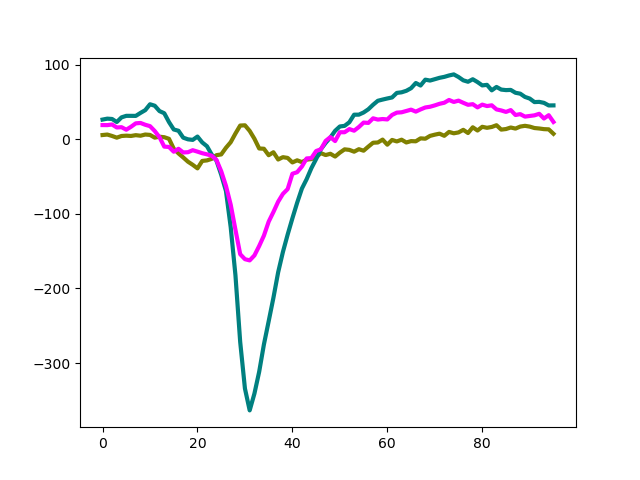
Compute unit maximum channel¶
In a similar way, one can get the recording channel with the ‘extremum’ signal (minimum or maximum). The code:get_template_extremum_channel outputs a dictionary unit_ids as keys and channel_ids as values:
extremum_channels_ids = st.get_template_extremum_channel(we, peak_sign='neg')
print(extremum_channels_ids)
Out:
{'#0': '6', '#1': '10', '#2': '20', '#3': '3', '#4': '14', '#5': '12', '#6': '25', '#7': '22', '#8': '23', '#9': '7'}
Compute principal components (aka PCs)¶
Computing PCA scores for each waveforms is very common for many applications, including unsupervised validation of the spike sorting performance.
- There are different ways to compute PC scores from waveforms:
“concatenated”: all waveforms are concatenated and a single PCA model is computed (channel information is lost)
“by_channel_global”: PCA is computed from a subset of all waveforms and applied independently on each channel
“by_channel_local”: PCA is computed and applied to each channel separately
In SI, we can compute PC scores with the compute_principal_components function
(which returns a WaveformPrincipalComponent object).
The pc scores for a unit are retrieved with the get_components function and
the shape of the pc scores is (n_spikes, n_components, n_channels).
Here, we compute PC scores and plot the first and second components of channel 8:
pc = st.compute_principal_components(we, load_if_exists=True,
n_components=3, mode='by_channel_local')
print(pc)
fig, ax = plt.subplots()
for i, unit_id in enumerate(sorting.unit_ids[:3]):
comp = pc.get_components(unit_id)
print(comp.shape)
color = colors[i]
ax.scatter(comp[:, 0, 8], comp[:, 1, 8], color=color)
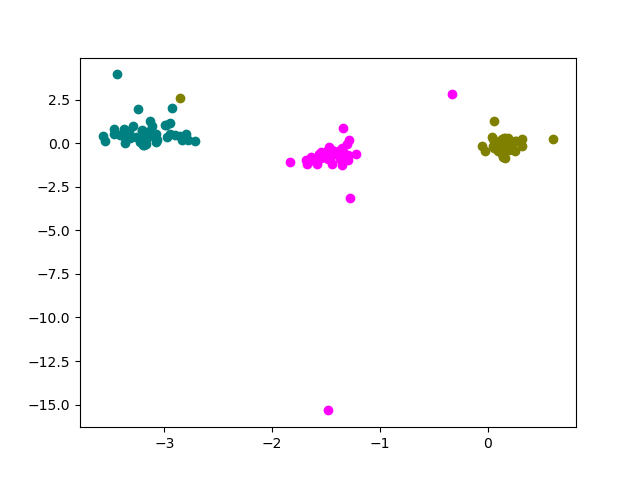
Out:
WaveformPrincipalComponent: 32 channels - 1 segments
mode:by_channel_local n_components:3
(53, 3, 32)
(50, 3, 32)
(43, 3, 32)
Note that PC scores for all units can be retrieved at once with the get_all_components() function:
all_labels, all_components = pc.get_all_components()
print(all_labels[:40])
print(all_labels.shape)
print(all_components.shape)
cmap = plt.get_cmap('Dark2', len(sorting.unit_ids))
fig, ax = plt.subplots()
for i, unit_id in enumerate(sorting.unit_ids):
mask = all_labels == unit_id
comp = all_components[mask, :, :]
ax.scatter(comp[:, 0, 8], comp[:, 1, 8], color=cmap(i))
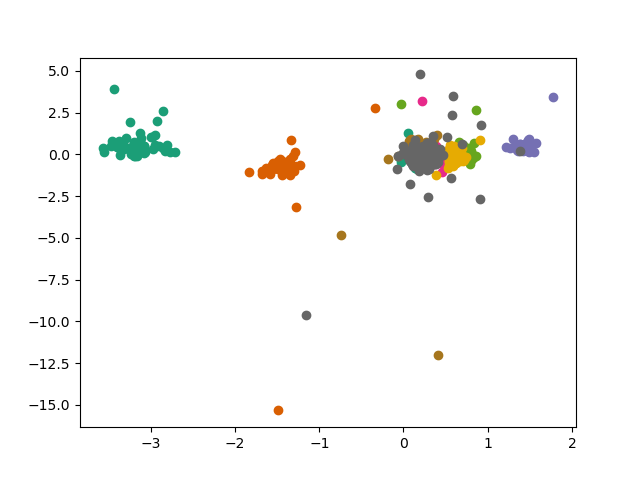
Out:
['#0' '#0' '#0' '#0' '#0' '#0' '#0' '#0' '#0' '#0' '#0' '#0' '#0' '#0'
'#0' '#0' '#0' '#0' '#0' '#0' '#0' '#0' '#0' '#0' '#0' '#0' '#0' '#0'
'#0' '#0' '#0' '#0' '#0' '#0' '#0' '#0' '#0' '#0' '#0' '#0']
(745,)
(745, 3, 32)
- Export sorted data to Phy for manual curation
@alessio : please remove this cell when you read it export_to_phy is not anymore in toolkit but in exporter
We won’t make a tutorial for it because it is super slow. Only statics docs. Finally, it is common to visualize and manually curate the data after spike sorting. In order to do so, we interface with the Phy GUI (https://phy-contrib.readthedocs.io/en/latest/template-gui/).
First, we need to export the data to the phy format:
# output_folder = 'mearec_exported_to_phy'
# st.export_to_phy(we,
# compute_pc_features=False, compute_amplitudes=True,
# remove_if_exists=True)
To run phy you can then run (from terminal):
phy template-gui mearec_exported_to_phy/params.py
Or from a notebook: !phy template-gui mearec_exported_to_phy/params.py
After manual curation you can load back the curated data using the PhySortingExtractor:
plt.show()
Total running time of the script: ( 0 minutes 2.117 seconds)